A highly collaborative team of scientists from ShanghaiTech University, The Hebrew University of Jerusalem, University of California, San Francisco and University of Southern California have developed a novel method called Bayesian metamodeling to integrate a collection of heterogeneous input models for the comprehensive modelling of the intact biological cells. The team provides a proof-of-principle metamodel of glucose-stimulated insulin secretion by human pancreatic β-cells. The research was published in the prestigious journal PNAS, available online on August 31, 2021, entitled ‘Bayesian metamodeling of complex biological systems across varying representations’.
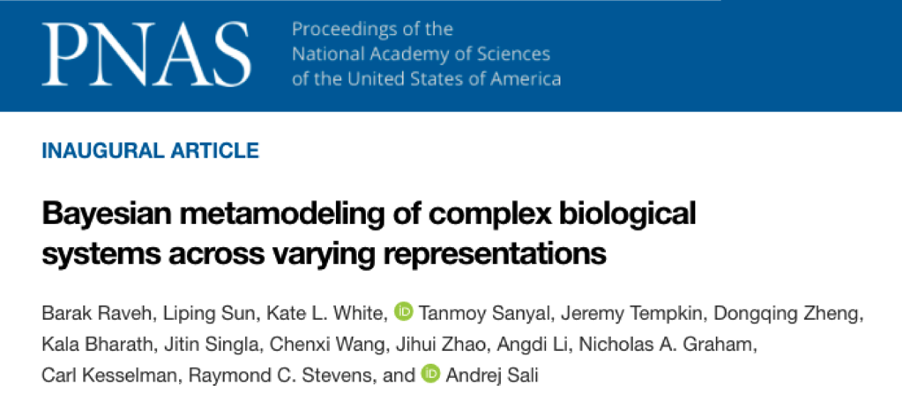
The initiation of building a spatiotemporal multi-scale whole cell model goes way back to the iHuman Forum in November 2017. Professor Andrej Sali drafted the opportunities and challenges in whole cell modelling based on the discussions in that forum, which was later published in Cell (https://pubmed.ncbi.nlm.nih.gov/29570991/). Since then, teams from different institutes have spent great efforts from experimentally mapping the cell to computationally modelling the subcellular organization, dynamics and signalling pathways to learn different aspects of β-cell biology. To divide-and-conquer such a large problem of modelling numerous aspects of the cell into computing a number of smaller models of different types, Bayesian metamodeling is developed to assemble these models into a complete map of the cell (Figure 1).
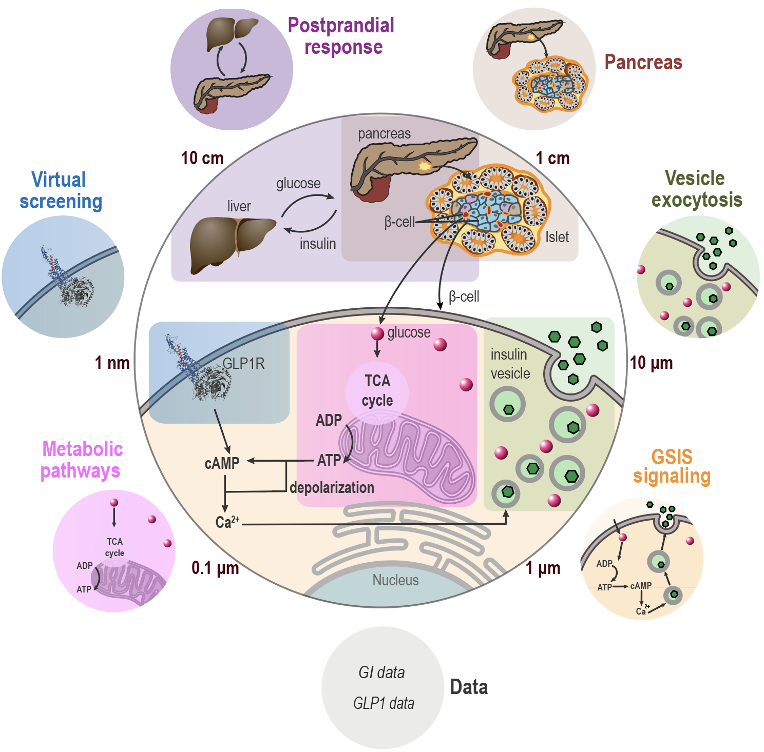
The laboratory at iHuman has been focused on developing and applying integrative modelling for a combined experimental and computational effort on mapping macromolecular assemblies and the whole cell. The lab has two major research directions, one is integrative modelling of the structural dynamics of GPCR G-protein complexes, and the other is integrative modelling and Bayesian metamodeling of the whole cell. Bayesian metamodeling proceeds along three steps (Figure 2): 1) converting heterogeneous input models into a standardized statistical representation relying on probabilistic graphical models, 2) coupling by modelling their mutual relations with the physical world, and 3) finally harmonizing with respect to each other. It often produces a more accurate, precise, and complete model that contextualizes input models as well as resolves conflicting information.
The research is a collaborative effort of scientists from four institutions. Founding Director Raymond Stevens, Distinguished Adjunct Professor Andrej Sali, Postdoctoral researchers Liping Sun, Jihui Zhao and Ph.D. students Chenxi Wang, Angdi Li are from the iHuman Institute at ShanghaiTech University. Beyond the iHuman team, Barak Raveh is from The Hebrew University of Jerusalem. Tanmoy Sanyal, Kala Bharath and Andrej Sali are from University of California, San Francisco. Kate White, Dongqing Zheng, Jitin Singla, Nicholas Graham, and Carl Kesselman are from University of Southern California.

