SHANGHAI, CHINA – April 30, 2021 – A team of scientists from ShanghaiTech University, Tianjin University, and The Agency for Science, Technology and Research (A*STAR)/ University of Illinois at Urbana-Champaign (UIUC) have solved a long-standing mystery of Z genome that comprises entirely non-canonical Z-T base pairs instead of canonical A-T base pairs in bacteriophages (Figure 1). They discovered the key enzymes responsible for the synthesis of this non-canonical base pair in dozens of globally widespread phages and further verified the Z genome in one of these phages. This discovery enables the synthesis of Z-DNA on a large scale for a diverse range of applications ranging from phage therapy to synthetic biology to DNA storage. The research is published in Science, available online on April 30, 2021.

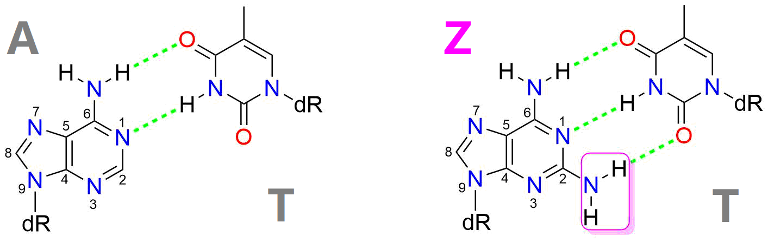
Figure 1. Z-T base pair has three hydrogen bonds, while A-T base pair only has two.
The laboratory of Suwen Zhao at iHuman Institute and School of Life Science and Technology, ShanghaiTech University, has been focused on developing computational methods to study protein sequence-structure-function relationship. The lab has two major research directions, one is GPCR mechanism study and ligand discovery, and the other is discovery of new enzyme functions, metabolic pathways, and metabolites. Suwen Zhao and the team have developed a series of computational methods to improve the accuracy and/or throughput of assigning reliable functions to enzymes discovered in various bacterial or viral genomes.
The laboratory of Yan Zhang at School of Pharmaceutical Science and Technology, Tianjin University has expertise in characterizing new enzymes and pathways through a combination of bioinformatic, genetic, biochemical and biophysical methods.
The laboratory of Huimin Zhao at A*STAR/UIUC develops and applies synthetic biology, machine learning, and laboratory automation tools to engineer functionally improved or novel proteins, pathways, and genomes for biotechnological and biomedical applications.
The team formed by researchers from these three labs is interested in predicting and validating functions of enzymes involved in phage DNA modifications. Phage DNA is often modified to evade digestion of host bacterial restriction enzymes. Among dozens of naturally occurring DNA modifications, most can be found in bacteriophages. Diamopurine (Z) is one of such cases. It was discovered in the genomic DNA of cyanophage S-2L more than 40 years ago. It replaces adenine (A) in the phage DNA entirely and forms three hydrogen bonds with thymine (T). However, the enzymes required for Z genome synthesis are not known.
The team applied in silico methods such as sequence similarity network analysis, protein structure prediction, genome context analysis, and enzyme mechanism study, combined with in vitro and in vivo experimental methods including various enzymatic assays, HPLC, LC-MS/MS and nanopore sequencing, to characterize the biosynthetic pathway responsible for Z genome synthesis in phages (Figure 2) and validate the Z genome in Acinetobacter phage SH-Ab 15497.
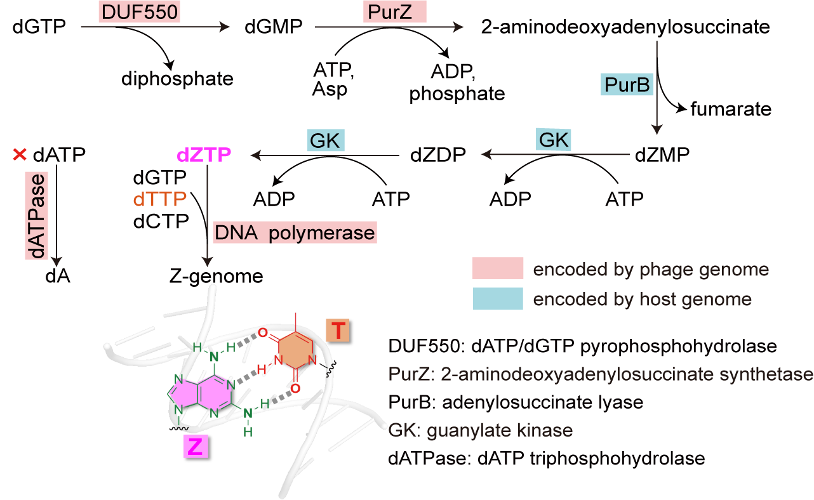
Figure 2. The characterized biosynthetic pathway of Z-genome in phage.
The research is a collaborative effort of scientists from six institutions. Ph.D. students Xuexia Xu, Yu Cheng and Yu Guo are from Suwen Zhao’s lab at iHuman Institute and School of Life Science and Technology, and Zhangyue Song is from the biomedical big data core in SIAIS. Beyond the ShanghaiTech team, Yan Zhou, Zhi Li, Yan Gao, and Yan Zhang are from Tianjin University; Yifeng Wei, Ee Lui Ang, and Huimin Zhao are from A*STAR/UIUC; Ivan Khudyakov, the discover of cyanophage S-2L, who provided valuable suggestions for the project, is from All-Russian Research Institute for Agricultural Microbiology, Saint Petersburg; and Fuli Liu and Ping He, who provided the Z-DNA of Acinetobacter phage SH-Ab 15497, are from Shanghai Jiao Tong University. The cloning core in iHuman, the biomedical big data core and analytical chemistry core in SIAIS also provided assistance.

