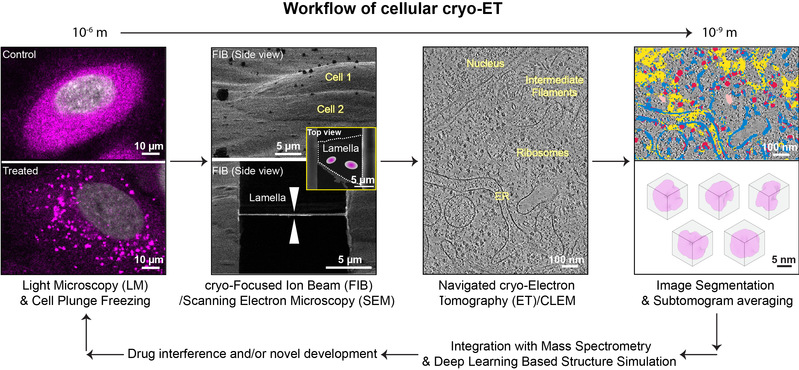
近年发展起来的冷冻聚焦离子束减薄技术打开了直接观察细胞生理状态下大分子互作的“窗口”。联合冷冻电子断层扫描技术、以及冷冻光电镜联用技术,课题组利用跨尺度方法研究细胞原位的结构功能事件,聚焦癌症这一日益严重的全球性健康问题。由于许多肿瘤相关的蛋白因子可以在细胞内形成微米尺度的凝聚体(通过相分离),参与到信号传导、代谢等关键事件,课题组将研究肿瘤发生及肿瘤免疫过程中的蛋白质凝聚体的组装模式。并将通过整合定量质谱学、人工智能辅助的复合物结构模拟、以及高分辨结构信息等解释其形成的分子机制。具体研究方向包括环状GMP-AMP合成酶凝聚体、免疫突触、以及肿瘤细胞代谢酶体等在细胞原位的高级组装模式,最终为肿瘤治疗药物设计提供新的思路。
1. Zhang, Xiaojie*#; Mahamid, Julia#; .Protocol for subtomogram averaging of helical filaments in cryo-electron tomography.STAR PROTOCOLS. Sep 2024.
2. Zhang, Xiaojie*; Sridharan, Sindhuja*; Zagoriy, Ievgeniia; Eugster Oegema, Christina; Ching, Cyan; Pflaesterer, Tim; Fung, Herman K.H.; Becher, Isabelle; Poser, Ina; Mu ̈ ller, Christoph W.; Hyman, Anthony A.; Savitski, Mikhail M.#; Mahamid, Julia#; .Molecular mechanisms of stress-induced reactivation in mumps virus condensates.CELL. Apr 2023.
3. Tollervey, Fergus*; Zhang, Xiaojie; Bose, Mainak; Sachweh, Jenny; Woodruff, Jeffrey B.; Franzmann, Titus M.; Mahamid, Julia#; .Cryo-Electron Tomography of Reconstituted Biomolecular Condensates.METHODS IN MOLECULAR BIOLOGY. Oct 2022.
4. Klumpe, Sven*; Fung, Herman KH*; Goetz, Sara K*; Zagoriy, Ievgeniia; Hampoelz, Bernhard; Zhang, Xiaojie; Erdmann, Philipp S; Baumbach, Janina; Müller, Christoph W; Beck, Martin; Plitzko, Jürgen M#; Mahamid, Julia#; .A modular platform for automated cryo-FIB workflows.ELIFE. Dec 2021.
5. Jawerth, Louise*; Fischer-Friedrich, Elisabeth; Saha, Suropriya; Wang, Jie; Franzmann, Titus; Zhang, Xiaojie; Sachweh, Jenny; Ruer, Martine; Ijavi, Mahdiye; Saha, Shambaditya; Mahamid, Julia; Hyman, Anthony A.#; Jülicher, Frank#; .Protein condensates as aging Maxwell fluids.SCIENCE. Dec 2020.
6. Allegretti, Matteo*; Zimmerli, Christian E.*; Rantos, Vasileios; Wilfling, Florian; Ronchi, Paolo; Fung, HermanK.H.; Lee, Chia Wei; Hagen, Wim; Turoňová, Beata; Karius, Kai; Börmel, Mandy; Zhang, Xiaojie; Müller, Christoph W.; Schwab, Yannick; Mahamid, Julia; Pfander, Boris#; Kosinski, Jan#; Beck, Martin#; .In-cell architecture of the nuclear pore and snapshots of its turnover.NATURE. Oct 2020.
7. Zhang, Xiaojie*; Mahamid, Julia#; .Addressing the challenge of in situ structural studies of RNP granules in light of emerging opportunities.CURRENT OPINION IN STRUCTURAL BIOLOGY. Jul 2020.
8. Guille ́ n-Boixet, Jordina*; Kopach, Andrii*; Holehouse, Alex S.; Wittmann, Sina; Jahnel, Marcus; Schlu ̈ ßler, Raimund; Kim, Kyoohyun; Trussina, Irmela R.E.A.; Wang, Jie; Mateju, Daniel; Poser, Ina; Maharana, Shovamayee; Ruer-Gruß, Martine; Richter, Doris; Zhang, Xiaojie; Chang, Young-Tae; Guck, Jochen; Honigmann, Alf; Mahamid, Julia; Hyman, Anthony A.; Pappu, Rohit V.; Alberti, Simon#; Franzmann, Titus M.; .RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation.CELL. Apr 2020.
9. Wang, Jie*; Choi, Jeong-Mo; Holehouse, Alex S.; Lee, Hyun O.; Zhang, Xiaojie; Jahnel, Marcus; Maharana, Shovamayee; Lemaitre, Re ́ gis; Pozniakovsky, Andrei; Drechsel, David; Poser, Ina; Pappu, Rohit V.; Alberti, Simon#; Hyman, Anthony A.#; .A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins.CELL. Jul 2018.
10. Negri, Ana*; Javidnia, Prisca*; Mu, Ran*; Zhang, Xiaojie*; Vendome, Jeremie; Gold, Ben; Roberts, Julia; Barman, Dipti; Ioerger, Thomas; Sacchettini, James C.; Jiang, Xiuju; Burns-Huang, Kristin; Warrier, Thulasi; Ling, Yan; Warren, J. David; Oren, Deena A.; Beuming, Thijs; Wang, Hongyao; Wu, Jie; Li, Haitao; Rhee, Kyu Y.; Nathan, Carl F.; Liu, Gang; Somersan-Karakaya, Selin#; .Identification of a Mycothiol-Dependent Nitroreductase from Mycobacterium tuberculosis.ACS INFECTIOUS DISEASES. Feb 2018.
11. Zhao, Dan*; Zhang, Xiaojie; Guan, Haipeng; Xiong, Xiaozhe; Shi, Xiaomeng; Deng, Haiteng; Li, Haitao#; .The BAH domain of BAHD1 is a histone H3K27me3 reader.PROTEIN & CELL. Mar 2016.
12. Zhang, Xiaojie*; Zhao, Dan; Xiong, Xiaozhe; He, Zhimin; Li, Haitao#; .Multifaceted Histone H3 Methylation and Phosphorylation Readout by the Plant Homeodomain Finger of Human Nuclear Antigen Sp100C.JOURNAL OF BIOLOGICAL CHEMISTRY. Apr 2016.










