Predicting the full-length structures of multi-domain GPCRs
Discovering ligands for GPCRs and other proteins in cell signaling pathways
My research group aim to develop and apply methods to gain understanding of protein sequence-structure-function relationship. We develop and apply methods in blue to address this problem. We work on enzymes and transmembrane receptors.
By analyzing the conformational changes of receptor structures, we discovered a pathway comprising of 34 residue pairs and 35 residues, exhibiting a common activation mechanism of class A receptors.

Zhou Q, Yang D, Wu M, Guo Y, Guo W, Zhong L, Cai X, Dai A, Jang W, Shakhnovich EI, Liu ZJ, Stevens RC, Lambert NA, Babu MM*, Wang MW*, Zhao S*.Common activation mechanism of class A GPCRs. eLife. 2019 Dec 19;8. pii: e50279. doi: 10.7554/eLife.50279
The Z genome consists of four nucleosides, ZTCG, of which the biosynthetic pathway was previously unknown. Through bioinformatic analysis, we have identified a multi-enzyme system responsible for the synthesis of Z nucleosides and the Z genome, and the functions of these enzymes have been validated in vitro and in vivo. Our study shows that Z-genome phages have a global distribution and that these phages can infect bacteria of the phyla Cyanobacteria, Actinomycetes and Proteobacteria, so it is likely that organisms with the ZTCG genome have existed on Earth in parallel with organisms with the ATCG genome since at least 3.5 billion years ago. The elucidation of the Z genome synthesis pathway will also advance research in Z genome-related synthetic biology.

Zhou Y.; Xu X.; Wei Y.; Cheng Y.; Guo Y.; Khudyakov I.; Liu F.; He P.; Song Z.; Li Z.; Gao Y.; Ang E. L.; Zhao H.*; Zhang Y.*; Zhao S.*, A widespread pathway for substitution of adenine by diaminopurine in phage genomes. Science, 2021 April 30, 372(6541):512-516. doi:10.1126/science.abe4882.
We built a high-quality dataset (LigBoundConf) of 8145 ligand-bound conformations from PDB crystal structures and calculated LCSE in the dataset. Our work provides a comprehensive profile of LCSE for ligands in PDB, which could help ligand conformation generation, ligand docking pose evaluation, and lead optimization.

Tong J, Zhao S*.Large-Scale Analysis of Bioactive Ligand Conformational Strain Energy by Ab Initio Calculation.J Chem Inf Model. 2021 Mar 22;61(3):1180-1192. doi: 10.1021/acs.jcim.0c01197.
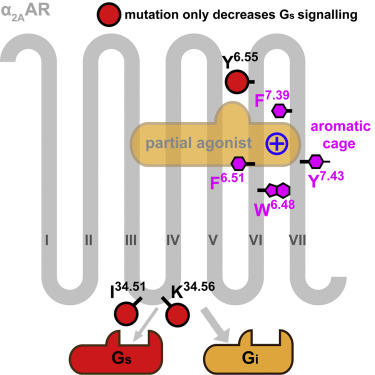
We have determined the partial agonist and antagonist-bound α2A crystal structures, and compared to the structure of partial agonist-bound, the full agonist-bound forms a hydrogen bond withY3946.55. We found that ICL2 plays an important role in the recruitment of Gs to the partial agonist-bound α2A. In contrast to the β Adrenergic, F4127.39 is critical at the adrenergic α2A.
we developed a nhmmer-based olfactory receptor annotation tool Genome2OR. 765,248 olfactory receptor genes were annotated, with 404,426 functional genes and 360,822 pseudogenes. Based on the annotation data, we built a database called Chordata Olfactory Receptor Database (CORD) for archiving, analysing and disseminating the data.
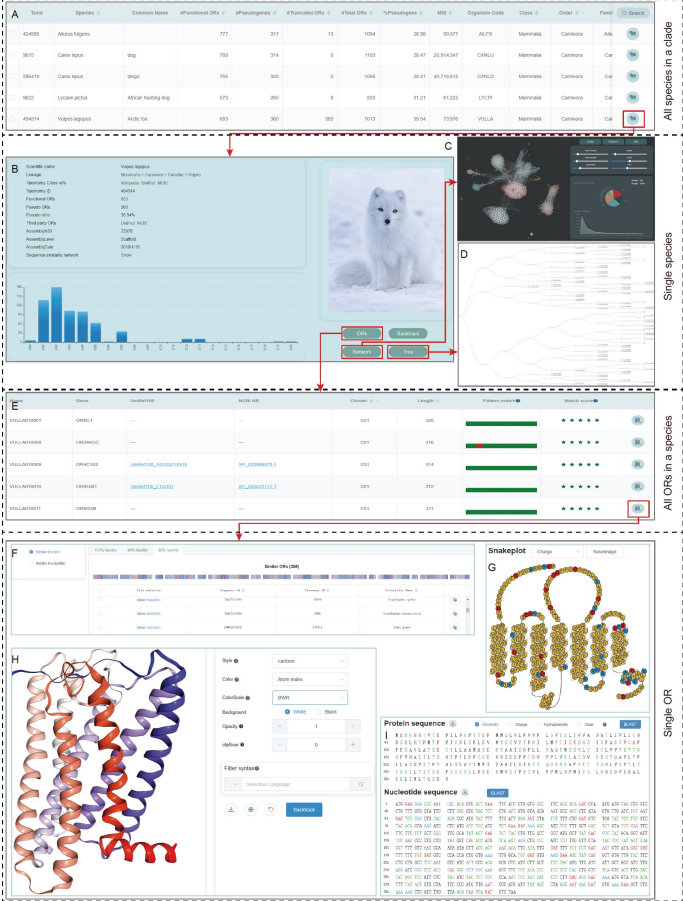
Han W, Wu Y, Zeng L, Zhao S*. Building the Chordata Olfactory Receptor Database using more than 400,000 receptors annotated by Genome2OR.Sci China Life Sci. 2022 Jun 10. doi: 10.1007/s11427-021-2081-6.
GPCR Allosteric Modulator Discovery. Wu, Yiran; Tong, Jiahui; Ding, Kang; Zhou, Qingtong; Zhao, Suwen. Book Chapter in Protein Allostery in Drug Discovery, Advances in Experimental Medicine and Biology. 1163:225-251

















