课题组主要致力于开发和应用各种计算化学和计算生物学方法来研究蛋白质序列-结构-功能关系关系。
研究方向:
(1)膜蛋白的药物研发与机理研究,
(2)酶的功能预测、机理研究以及代谢通路预测。
课题组开发的网站:
脊索动物嗅觉受体数据库 Chordata Olfactory Receptor Database (CORD)
https://cord.ihuman.shanghaitech.edu.cn
我们通过残基接触打分(Residue-Residue Contact Score, RRCS)分析了A家族GPCR 200多个结构残基层面的构象变化,发现A家族GPCR共享一个由34个氨基酸残基对(35个残基)构成的激活通路,揭示了A家族GPCR的共同激活机制。

Zhou Q, Yang D, Wu M, Guo Y, Guo W, Zhong L, Cai X, Dai A, Jang W, Shakhnovich EI, Liu ZJ, Stevens RC, Lambert NA, Babu MM*, Wang MW*, Zhao S*.Common activation mechanism of class A GPCRs. eLife. 2019 Dec 19;8. pii: e50279. doi: 10.7554/eLife.50279
Z基因组由四种核苷,ZTCG组成,其中的生物合成途径此前是未知的。通过生物信息学分析,我们确定了一个负责Z基因组合成的多酶系统,这些酶的功能已经在体外和体内得到验证。我们的研究表明,Z基因组噬菌体具有遍布全球,这些噬菌体可以感染蓝藻门、放线菌门和变形菌门的细菌。因此推测至少在35亿年前,具有ZTCG基因组的生物体很可能已经与具有ATCG基因组的生物体同时存在于地球上。Z基因组合成通路的阐明也将推动Z基因组相关的合成生物学的研究。

Zhou Y.; Xu X.; Wei Y.; Cheng Y.; Guo Y.; Khudyakov I.; Liu F.; He P.; Song Z.; Li Z.; Gao Y.; Ang E. L.; Zhao H.*; Zhang Y.*; Zhao S.*, A widespread pathway for substitution of adenine by diaminopurine in phage genomes. Science, 2021 April 30, 372(6541):512-516. doi:10.1126/science.abe4882.
我们从PDB晶体结构出发,筛选构建了一个含有8145个配体结合构象的高质量数据集。针对该数据集,我们计算了配体的构象张力能(Ligand conformational strain energy, LCSE),为PDB中配体的LCSE提供了一个全面的画像,这将有助于配体构象生成,配体结合模式评估,以及通过减少张力能来进行先导化合物优化。

Tong J, Zhao S*.Large-Scale Analysis of Bioactive Ligand Conformational Strain Energy by Ab Initio Calculation.J Chem Inf Model. 2021 Mar 22;61(3):1180-1192. doi: 10.1021/acs.jcim.0c01197.
我们分别解析了结合部分激动剂以及拮抗剂的肾上腺素受体α2A晶体结构,同结合部分激动剂的结构比较,全激动剂会与Y3946.55形成氢键。我们发现在结合部分激动剂的肾上腺素受体α2A招募Gs的过程中ICL2扮演重要角色。同β肾上腺素受体相比, F4127.39在肾上腺素受体α2A至关重要。

我们基于nhmmer开发了一个嗅觉受体注释工具Genome2OR,最终从1608个脊索动物基因组中注释出765248个嗅觉受体,其中有404426个功能基因以及360822个假基因。在注释数据的基础上,我们建立了一个名为脊索动物嗅觉受体数据库(Chordata Olfactory Receptor Database,CORD)的数据库,用于存档、分析和传播数据。
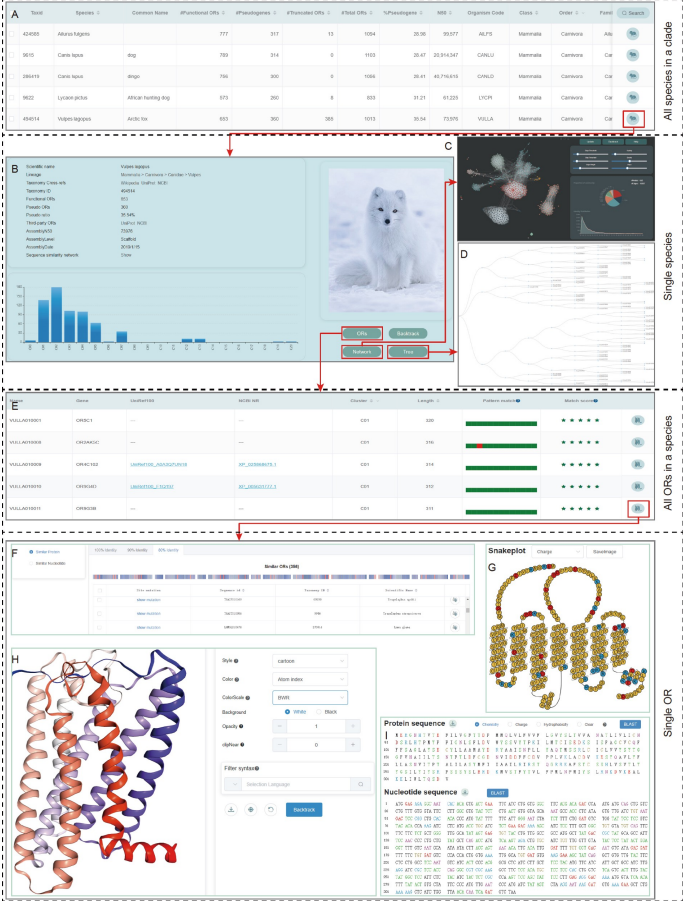
Han W, Wu Y, Zeng L, Zhao S*. Building the Chordata Olfactory Receptor Database using more than 400,000 receptors annotated by Genome2OR.Sci China Life Sci. 2022 Jun 10. doi: 10.1007/s11427-021-2081-6.
1. Building the Chordata Olfactory Receptor Database using more than 400,000 receptors annotated by Genome2OR.
Han W, Wu Y, Zeng L, Zhao S*. Sci China Life Sci. 2022 Jun 10. doi: 10.1007/s11427-021-2081-6.
2. A widespread pathway for substitution of adenine by diaminopurine in phage genomes.
Zhou Y#, Xu X#, Wei Y, Cheng Y, Guo Y, Khudyakov I, Liu F, He P, Song Z, Li Z, Gao Y, Ang E. L, Zhao H*, Zhang Y*, Zhao S*. Science. 2021 Apr 30;372(6541):512-516. doi: 10.1126/science.abe4882.
3. Large-Scale Analysis of Bioactive Ligand Conformational Strain Energy by Ab Initio Calculation.
Tong J, Zhao S*. J Chem Inf Model. 2021 Mar 22;61(3):1180-1192. doi: 10.1021/acs.jcim.0c01197.
4. Common activation mechanism of class A GPCRs.
Zhou Q, Yang D, Wu M, Guo Y, Guo W, Zhong L, Cai X, Dai A, Jang W, Shakhnovich EI, Liu ZJ, Stevens RC, Lambert NA, Babu MM*, Wang MW*, Zhao S*. eLife. 2019 Dec 19;8. pii: e50279. doi: 10.7554/eLife.50279.
5. Structural basis of the diversity of adrenergic receptors.
Qu L, Zhou Q, Xu Y, Guo Y, Chen X, Yao D, Han GW, Liu ZJ, Stevens RC, Zhong G*, Wu D*, Zhao S*. Cell Rep. 2019 Dec 3;29(10):2929-2935.e4. doi: 10.1016/j.celrep.2019.10.088.
6. Colocalization Strategy Unveils an Underside Binding Site in the Transmembrane Domain of Smoothened Receptor.
Zhou F, Ding K, Zhou Y, Liu Y, Liu X, Zhao F, Wu Y, Zhang X, Tan Q, Xu F, Tan W, Xiao Y*, Zhao S*, Tao H*. J Med Chem. 2019 Nov 14;62(21):9983-9989. doi: 10.1021/acs.jmedchem.9b00283.
7. Salvianolic acids from antithrombotic Traditional Chinese Medicine Danshen are antagonists of human P2Y1 and P2Y12 receptors.
Liu X, Gao Z, Wu Y, Stevens RC, Jacobson KA, Zhao S*. Sci Rep. 2018 May 24;8(1):8084. doi: 10.1038/s41598-018-26577-0.
8. Intrinsically disordered proteins link alternative splicing and post-translational modifications to complex cell signaling and regulation.
Zhou J, Zhao S*, Keith Dunker A*, J Mol Biol. 2018 Apr 4. pii: S0022-2836(18)30179-7. doi: 10.1016/j.jmb.2018.03.028.
9. Crystal structures of agonist-bound human cannabinoid receptor CB1.
Hua T, Vemuri K, Nikas S, Laprairie RB, Wu Y, Qu L, Pu M, Korde A, Shan J, Ho J. H, Han G. W, Ding K, Li X, Liu H, Hanson M. A, Zhao S*, Bohn L. M*, Makriyannis, A*, Stevens, R. C, Liu, Z. J*. Nature. 2017 Jul 27;547(7664):468-471. doi: 10.1038/nature23272.
10. Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Vetting MW#; Al-Obaidi N#; Zhao S#; San Francisco B#; Kim J, Wichelecki DJ, Bouvier JT, Solbiati JO, Vu H, Zhang X, Rodionov DA, Love JD, Hillerich BS, Seidel RD, Quinn RJ*, Osterman AL*, Cronan JE*, Jacobson MP*, Gerlt JA*, Almo SC*. Biochemistry. 2015 Jan 27;54(3):909-31. doi: 10.1021/bi501388y.
11. Prediction and characterization of enzymatic activities guided by sequence similarity and genome neighborhood networks.
Zhao S, Sakai A, Zhang X, Vetting MW, Kumar R, Hillerich B, San Francisco B, Solbiati J, Steves A, Brown S, Akiva E, Barber A, Seidel R. D, Babbitt PC, Almo SC*, Gerlt JA*, Jacobson MP*. eLife. 2014 Jun 30;3. doi: 10.7554/eLife.03275.
12. Discovery of new enzymes and metabolic pathways by using structure and genome context.
Zhao S, Kumar R, Sakai A, Vetting MW, Wood BM, Brown S, Bonanno JB, Hillerich BS, Seidel RD, Babbitt PC, Almo SC, Sweedler JV*, Gerlt JA*, Cronan JE*, Jacobson MP*. Nature. 2013 Oct 31;502(7473):698-702. doi: 10.1038/nature12576.
GPCR Allosteric Modulator Discovery. Wu, Yiran; Tong, Jiahui; Ding, Kang; Zhou, Qingtong; Zhao, Suwen. Book Chapter in Protein Allostery in Drug Discovery, Advances in Experimental Medicine and Biology. 1163:225-251

















